INsilico Medicine
PHARMA.AI
We explicitly strive to accelerate three areas of drug discovery and development:
Disease Target identification, Generation of Synthetic biology and Generation of Novel molecules data,
and Predicting Clinical trial outcomes
Our revolutionary drug discovery engine utilizes millions of data samples and multiple data types to discover signatures
of diseases and identify the most promising targets for billions of molecules that already exist or can be generated de novo with preferred sets of parameters
Disease Target identification, Generation of Synthetic biology and Generation of Novel molecules data,
and Predicting Clinical trial outcomes
Our revolutionary drug discovery engine utilizes millions of data samples and multiple data types to discover signatures
of diseases and identify the most promising targets for billions of molecules that already exist or can be generated de novo with preferred sets of parameters
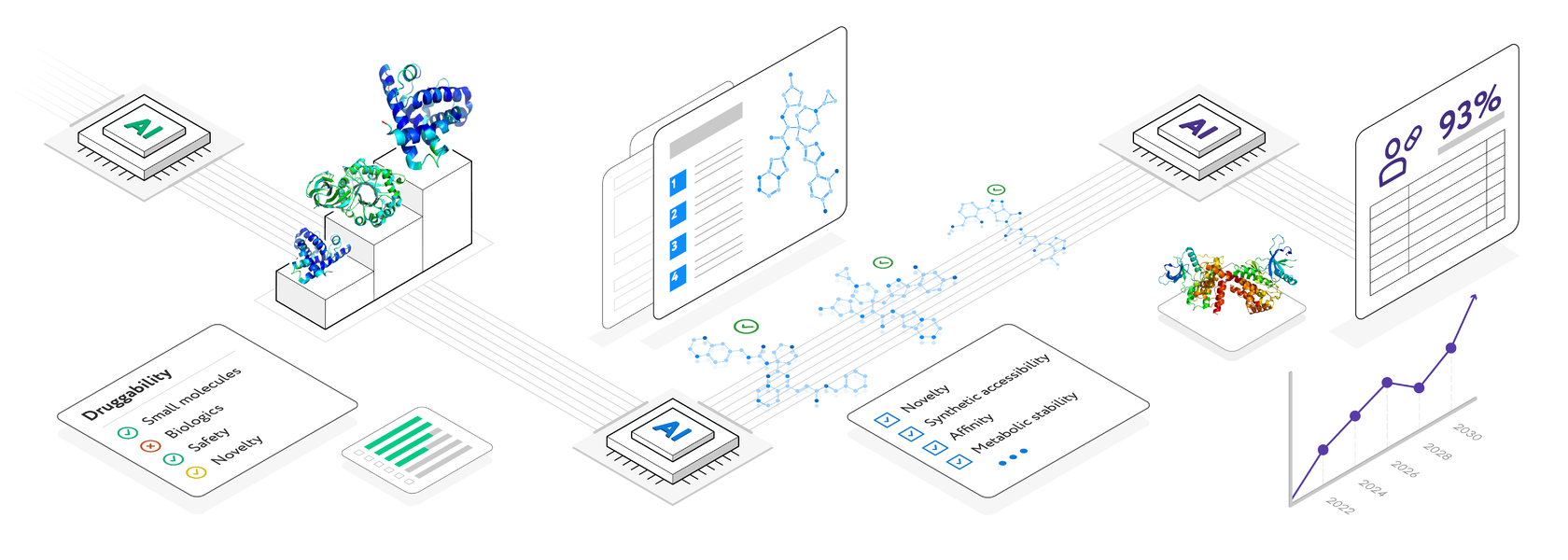
Enabling multi-omics target discovery and deep biology analysis engine to considerably reduce required time from several months to the span of just a few clicks
Find novel lead-like molecules in a week through this automated, machine learning de-novo drug design and scalable engineering platform
Predict clinical trials success rate, recognize the weak points in trial design, while adopting the best practices in the industry
Discover and Prioritize
Generate
Design and predict
Novel Targets
Novel Molecules
Clinical Trials

OMICs RESEARCH
AND TARGET ID
AND TARGET ID
OMICs data ANALYSIS
Access the full set of OMICs data generated by the scientific community so far. You do not need to spend your time trying to convert your data into an interpretable format or wait for a bioinformatician to do that for you — instead, you will find all the data already processed and uploaded in a uniform way, so you can focus on science and data interpretation.

Comprehensive
PATHWAY ANALYSIS
PATHWAY ANALYSIS
Pathway analysis is a crucial step toward a complete understanding of how data works. It converts a list of seemingly unrelated genes into a connected story based on dysregulated molecular processes. PandaOmics uses a proprietary pathway analysis approach called iPanda to infer pathway activation or inhibition. Results published in Nature communications in 2016 demonstrated the algorithm outperforming other pathway analysis tools.
Proof of concept
in Nature Communications
in Nature Communications

PRIORITIZATION
PandaOmics scoring approach is based on the combination of multiple scores derived from text and omics data associating genes with a disease of interest. This kind of approach allows us to unveil the hidden hypotheses that might not be obvious over common general knowledge or simple bioinformatics analysis. AI tools are extremely helpful for efficient target hypothesis generation. The overall scoring approach results in the ranked list of target hypothesis for a given disease (or disease subtype).
Proof of concept
in Nature Communications
in Nature Communications
Proof of concept
in ACS Publications
in ACS Publications

FILTERING
Once the gene list is prioritized according to the required strategy. Druggability filter uses traffic light logic and allows to filter target hypotheses according to their novelty, accessibility by small molecules and biologics and safety. Safety accounts for the involvement of the genes in toxicity related pathways and lethality of the corresponding gene knockout in mice. Additional filters include the protein and mRNA expression tissue specificity and protein class


OPTIMIZATION
Every new compound generated is annotated
with all the properties, including physico-chemical parameters, binding scores, drug-likeness features and mapped on vendors' catalogs and proprietary libraries for any similarity and novelty
with all the properties, including physico-chemical parameters, binding scores, drug-likeness features and mapped on vendors' catalogs and proprietary libraries for any similarity and novelty


PREDICTORS OF
CLINICAL TRIAL OUTCOMES
CLINICAL TRIAL OUTCOMES
FEATURE IMPACT
InClinico

NEWS




